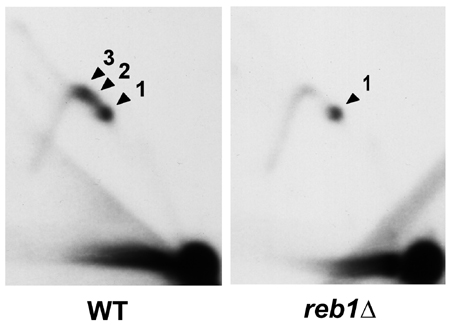
In eukaryotic cells, the deleterious effects of collision between transcription and replication are avoided by the presence of polar replication barriers. These barriers prevent replication complexes moving in the direction opposite to transcription to enter into transcriptional units. This is the case of the genes coding for ribosomal RNA (rDNA), where there is a temporal coincidence between transcription and replication. Our results indicate that this polar blockage is mediated by binding proteins that specifically recognize DNA sequences located in the barrier, arresting the replisome probably by inhibiting DNA helicase activity. In the mouse rDNA, replication seems to be blocked by the TTF-1 factor. This factor is also involved in the termination of rRNA transcription by RNA polymerase I. Therefore, it is a system in which the same cis-acting and trans-acting factors are able to arrest transcription approaching from upstream and replication approaching from downstream, preventing head-on collision of both machineries. We are characterizing this mechanism by using Schizosaccharomyces pombe as model.
A new type of mutations, known as dynamic mutations, is responsible for a series of human hereditary diseases. These mutations consist in the expansion of the copy number of trinucleotide repeats located in some genes. This expansion alters the expression of the affected gene. Our objective is to determine the role that DNA replication plays in the formation of these mutations. We have found that in simple model systems these trinucleotide repeats block replication complex movement in an orientation dependent manner. Our results indicate that replication slippage of the nascent strand takes place in the arrested replication forks leading to the expansion of the trinucleotide repeats.

