Group Leader/s
intro
Cytomotive proteins from the tubulin superfamily self-assemble forming dynamic filaments that are essential for DNA segregation, cell division, cytoplasmic organization and motility. These filaments translocate (treadmill) to perform their functions, even without motor proteins. They grow from one end while shortening from the other, by means of nucleotide hydrolysis and a polymerization-induced structural switch (see How protein filaments treadmill, Biophys J. 2020). Tubulin is the target of antitumor drugs that impair microtubule dynamics. FtsZ assembly guides bacterial cell division, which is a target for the discovery of new antibiotics needed to fight resistant pathogens.
Our work has focused on understanding how these protein assembly machines self-organize and work, how they evolved, and on targeting them with small molecules, employing complementary approaches at CIB and collaborating labs. Our group pioneered protein shape determination from synchrotron X-ray scattering with bead models and genetic algorithms. We have developed fluorescence polarization screening assays for antitumor inhibitors binding to the taxane site of microtubules, as well as fluorescent assays and cell-based methods to screen for GTP-replacing and allosteric antibacterial FtsZ inhibitors.
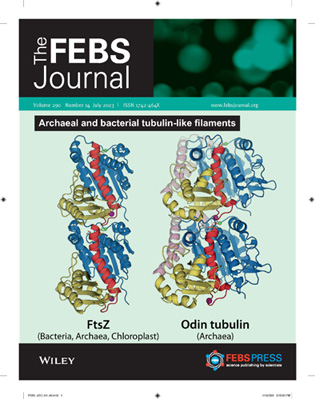
Following my formal retirement, I have pursued collaborative research interests at CIB as an emeritus scientist. This includes revealing the structural mechanisms of nucleotide hydrolysis and assembly dynamics of FtsZ from the leading pathogen Staphylococcus aureus, at atomic resolution (see Ruiz et al - PLOS Biology 2022), which bears implications for tubulin evolution (see Conserved GTPase mechanism in bacterial FtsZ and archaeal tubulin filaments, FEBS J 2023). I value interdisciplinary research and enjoy scientific discussions, biophysical methods and instruments.
Please click on research lines to see my timeline, former lab members and collaborators
direct web address: http://www.cib.csic.es/tubulinas
contact: j.m.andreu@cib.csic.es
Google profile: http://scholar.google.es/citations?hl=en&user=FyBdDrgAAAAJ&view_op=list_works&sortby=pubdate
TUBULIN AND FtsZ ASSEMBLY MACHINES.
Tubulin-like proteins spread in eukaryotes, bacteria, archaea, plasmids and viruses; they include microtubule αβ-tubulin, microtubule organizing γ-tubulin, primitive bacterial tubulin BtubA/B, bacterial cell division protein FtsZ, and the DNA-positioning TubZs codified by plasmid and phages, among other. They share a three-dimensional core structure, consisting of an N-terminal GTP binding domain and a GTPase activating domain connected by a central α-helix, but have divergent C-terminal secondary structures and disordered tails. Tubulins assemble head to tail into polar protofilaments with a 4 nm axial spacing between subunits that coalesce into different types of polymers, forming characteristic subcellular structures such as spindles or division rings. Assembly involves the formation of a subunit-subunit interface where the GTP-binding domain of one subunit in a protofilament interacts with the GTPase activation domain of the next subunit, which complements the GTP pocket and induces GTP hydrolysis, which triggers disassembly.
Tubulin and bacterial FtsZ each adopt two main conformations: the free unassembled proteins are in low self-association affinity, relaxed (R) structures, whereas the subunits in polymers (microtubules and FtsZ filaments) are hold in high self-association affinity, tense (T) forms. Switching from the R to the T state is coupled to the formation of a tight longitudinal association interface between consecutive subunits in the filament, rather than to the nucleotide state by itself. The assembly switch entails a rotation of the GTPase-activating domain with respect to the GTP-binding domain (FtsZ: open interdomain cleft = T vs closed cleft = R; Tubulin: straight subunits = T vs curved subunits = R). The local interactions made by the nucleotide gamma phosphate chemically enhance the affinity of the longitudinal association interface, rather than inducing large structural changes. Tubulin makes additional lateral contacts with neighbor protofilaments in microtubules.
The cooperative assembly of single-stranded FtsZ filaments and their treadmilling mechanism were puzzles whose solutions can also illuminate the dynamic mechanisms of the more complex cytomotive filaments. FtsZ monomers switching between low (R) and high affinity (T) conformations explains nucleated condensation polymerization of linear filaments, as well as the directional filament treadmilling upon GTP hydrolysis at the association interfaces. FtsZ filament treadmilling around the division ring guides peptidoglycan synthesis during bacterial division. Interestingly, the tubulin assembly switch may as well enable microtubule treadmillling and dynamic instability. Actin filament treadmilling appears to involve an analogous mechamism, in which G-actin may be identified as a R-state and F-actin as a T-state.



PC190723 GDP taxol GDP TubZ from botulinum cs-t phage
Ligand binding cavities in FtsZ (left) and beta-tubulin (right)


